Function and expression evaluation#
In this section, we will look at how we can extract data once we have solved our variational problem. To make this section concise, we will use functions with expressions that are already defined (not through a PDE), but through interpolation.
We start by creating a 3D mesh
from mpi4py import MPI
import numpy as np
import dolfinx
N = 2
mesh = dolfinx.mesh.create_box(
MPI.COMM_WORLD,
[np.array([0, 0, 0]), np.array([2, 1.3, 0.8])],
[N, N, N],
dolfinx.mesh.CellType.tetrahedron,
ghost_mode=dolfinx.mesh.GhostMode.shared_facet,
)
We start by considering a scalar function in a discontinuous space
V = dolfinx.fem.functionspace(mesh, ("DG", 2))
u = dolfinx.fem.Function(V)
Interpolation on a subset of cells#
We would like to interpolate the function
We start by locating the cells that satisfies this condition.
In Locating a subset of entities on a boundary we learnt how to use
dolfinx.mesh.locate_entities_boundary() to locate entities.
In this section we will use dolfinx.mesh.locate_entities()
which does the same, but for all entities in the mesh.
tdim = mesh.topology.dim
left_cells = dolfinx.mesh.locate_entities(mesh, tdim, lambda x: x[0] <= 1 + 1e-14)
right_cells = dolfinx.mesh.locate_entities(mesh, tdim, lambda x: x[0] >= 1 - 1e-14)
We can now interpolate the function onto each of these subsets
u.interpolate(lambda x: x[0] ** 2, cells0=left_cells)
u.interpolate(lambda x: x[1], cells0=right_cells)
Whenever we interpolate on sub-sets of cells, we need to scatter forward the values
u.x.scatter_forward()
2025-10-30 19:19:12.364 ( 0.790s) [ 7FEA15BF5140]vtkXOpenGLRenderWindow.:1458 WARN| bad X server connection. DISPLAY=
Evaluation at a point#
We want to evaluate the function at a point in the mesh that does not align with the nodes of the mesh. We do this through a sequence of steps:
As the mesh will be distributed on multiple processes, we have to determine which processes that has the point.
Then, for the processes that has the point, we need to figure out what cell the point is in.
Finally, we push this point back to the reference element, so that we can evaluate the basis functions, combine them with the coefficients on the given cell and push them forward to the physical space.
Step 1: Determine which processes that has the cell#
As looping through all the cells, and compute exact collisions is expensive,
we accelerate the search by using an axis-aligned bounding box tree.
This is a tree that recursively divides the mesh into smaller and smaller boxes, such that we can quickly
search through the tree to find the cells that might contain the point of interest.
bb_tree = dolfinx.geometry.bb_tree(mesh, mesh.topology.dim, padding=1e-10)
Note
Bounding boxes of other entities As seen below, we send in the topological dimension of the entities that we are interested in. This means that you can make a bounding box tree for facets, edges or vertices as well if it is needed.
Bounding boxes of subsets of entities
In many scenarios, we already have a notion about which entities we are interested in, we can create a bounding box tree of only these entities by sending in the entities as a list.
sub_bb_tree = dolfinx.geometry.bb_tree(mesh, mesh.topology.dim, entities=left_cells, padding=1e-10)
We can now send the points into dolfinx.geometry.compute_collisions_points()
to find the cells that contain the point.
Point collision in parallel
Note that each bounding box tree is local to the given process, and thus we can send in the same point to all processes, or unique points for each process.
points = np.array([[0.51, 0.32, 0], [1.3, 0.834, 0]], dtype=mesh.geometry.x.dtype)
potential_colliding_cells = dolfinx.geometry.compute_collisions_points(bb_tree, points)
potential_colliding_cells._cpp_object=<AdjacencyList> with 2 nodes
0: [25 13 17 10 21 29 ]
1: [4 14 8 16 22 9 ]
Output of compute point collisions
As we observe above, we get an AdjacencyList
out of the function compute_collisions_points.
This is a list of lists, where the \(ith\) list contains the indices of the cells that has bounding
boxes that collide with the \(i\)th point.
potential_colliding_cells.links(0)=array([25, 13, 17, 10, 21, 29], dtype=int32)
Step 2: Determine what cells the point is in#
Now that we have a reduced number of cells to more carefully examine, we can use the Gilbert–Johnson–Keerthi distance algorithm GJK. To find the cells whose convex hull contains the point.
Higher order meshes
As the GJK algorithm work on convex hulls, it is not 100 % accurate for higher order geometries (i.e. coordinate element is higher order, and the facets are curved). However, it is usually a sufficiently good enough approximation to be used in practice. For a more accurate algorithm, one could take all the points that satisfies the boundary box collision, pull them back to the reference element, and check that the resulting coordinate are inside the reference element. However, for non-affine grids this involves solving a non-linear problem (with a Newton type method).
To do this efficiently, we use dolfinx.geometry.compute_colliding_cells()
which takes in the output of compute_collisions_points and the points.
colliding_cells = dolfinx.geometry.compute_colliding_cells(mesh, potential_colliding_cells, points)
colliding_cells._cpp_object=<AdjacencyList> with 2 nodes
0: [10 ]
1: [8 ]
If a point is on an interface between two cells, we can get multiple cells that contain the point. Now, to prepare for step 3, we reduce the set of points and cells to those that are colliding on the current process.
points_on_proc = []
cells = []
for i, point in enumerate(points):
if len(colliding_cells.links(i)) > 0:
points_on_proc.append(point)
cells.append(colliding_cells.links(i)[0])
Step 3: Evaluate function at a point in the cell#
We now have on step left, which is to evaluate the function at the point in the cell.
As we saw above, this step involves quite a few operations.
Thankfully, these are all encapsulated in the function dolfinx.fem.Function.eval().
points_on_proc = np.array(points_on_proc, dtype=np.float64).reshape(-1, 3)
cells = np.array(cells, dtype=np.int32)
u_values = u.eval(points_on_proc, cells)
points_on_proc=array([[0.51 , 0.32 , 0. ],
[1.3 , 0.834, 0. ]])
u_values=array([[0.2601],
[0.834 ]])
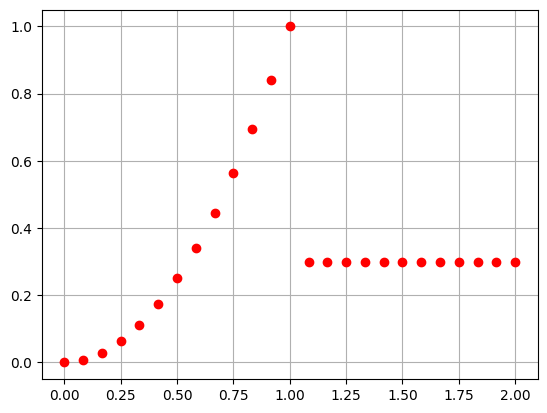
Evaluating along a line \(y=0.3, z=0.2\) and plot the result
Expand the dropdowns below to see the solution
Evaluate a UFL expression#
As we have seen in Generating code for assembling tensors we can generate code for evaluating integrals. We can also non-integrated UFL expressions at any point in the mesh. For this part of the tutorial, we will consider a blocked Lagrange space, with 3 components.
mesh = dolfinx.mesh.create_unit_cube(MPI.COMM_WORLD, 5, 5, 5, dolfinx.cpp.mesh.CellType.hexahedron)
V = dolfinx.fem.functionspace(mesh, ("Lagrange", 2, (3,)))
u = dolfinx.fem.Function(V)
def f(mod, x):
return x[2] ** 2, x[0] + x[1], x[1] * x[2]
u.interpolate(lambda x: f(np, x))
We will consider
We can write this in UFL as
import ufl
We can use dolfinx.fem.Expression to compile the evaluation of this expression
at a set of points in the reference element.
For instance, we can choose the point \((0.2, 0.3, 0.5)\) which is in the reference
hexahedron.
points = np.array([[0.2, 0.3, 0.5]], dtype=np.float64)
expr = dolfinx.fem.Expression(curl_u, points)
We can now evaluate the expression at this point in any cell in the mesh We pick two random cells on the process
num_cells_local = mesh.topology.index_map(mesh.topology.dim).size_local
cells = np.random.randint(0, num_cells_local, 2, dtype=np.int32)
values = expr.eval(mesh, cells)
We can inspect what the coordinates in the physical cell is by using the
same strategy of ufl.SpatialCoordinate(mesh)
x_expr = dolfinx.fem.Expression(ufl.SpatialCoordinate(mesh), points)
coords = x_expr.eval(mesh, cells)
values=array([[[0.7, 1.4, 1. ]],
[[0.1, 0.2, 1. ]]])
coords=array([[[0.64, 0.86, 0.7 ]],
[[0.24, 0.86, 0.1 ]]])
What points in the reference cells could be interesting to evaluate at?
In theory any point could be interesting to evaluate at, but usually evaluating at a set of quadrature points (for debugging) or at the interpolation points of a finite element.
Interpolation of a UFL expression#
We can for instance interpolate the gradient of a function into another function space. We will be inspired by the De Rahm complex And interpolate the gradient of a \(H^1\) function into \(H^{k-1}(curl)\)
Q = dolfinx.fem.functionspace(mesh, ("Lagrange", 2))
q = dolfinx.fem.Function(Q)
def f(x):
return x[0] ** 2 + 2 * x[1] ** 2 + x[1] * x[2]
q.interpolate(f)
grad_q = ufl.grad(q)
P = dolfinx.fem.functionspace(mesh, ("N1curl", 2))
p = dolfinx.fem.Function(P)
grad_expr = dolfinx.fem.Expression(grad_q, P.element.interpolation_points)
p.interpolate(grad_expr)
def grad_f(x):
return (2 * x[0], 4 * x[1] + x[2], x[1])
x = ufl.SpatialCoordinate(mesh)
f_ex = ufl.as_vector(grad_f(x))
L2_error = dolfinx.fem.form(ufl.inner(p - f_ex, p - f_ex) * ufl.dx)
local_error = dolfinx.fem.assemble_scalar(L2_error)
global_error = np.sqrt(mesh.comm.allreduce(local_error, op=MPI.SUM))
global_error=np.float64(2.36718250466125e-14)
Expression evaluation on facets#
Another neat feature for coupling to other codes it that we can evaluate
expressions on facets. This could for instance involve the
ufl.FacetNormal, which represents the normal point out of any facet of the cell.
We can for instance consider the heat flux on the boundary of a domain.
Integration entities#
Until now, we have represented the different entities of the domain with a given local index. This was the case for both cells, facets, edges and vertices. However, to be able to define the side of a facet we would like to evaluate an expression, we need to associate it with a cell. We do this by looking at all the facets associated with the cell, and then find its local index.
Integration entity
There are three distinct integration entities in FEniCS:
For cell integrals: The cell index itself is the integration entity
For exterior facet integrals: The cell index and the local facet index is the integration entity as a tuple
(cell, local_facet)For interior facet integrals: A tuple consisting of the
(cell, local_facet)for both cells connected to the facet, i.e.(cell_0, local_facet_0, cell_1, local_facet_1)
We will illustrate this below, by first finding all facets on one of the boundaries of the mesh
tdim = mesh.topology.dim
set_of_facets = dolfinx.mesh.locate_entities_boundary(mesh, tdim - 1, lambda x: np.isclose(x[1], 1))
Next, we create the appropriate connectivity in the mesh to be able to find the local index
mesh.topology.create_connectivity(tdim - 1, tdim)
f_to_c = mesh.topology.connectivity(tdim - 1, tdim)
mesh.topology.create_connectivity(tdim, tdim - 1)
c_to_f = mesh.topology.connectivity(tdim, tdim - 1)
Next, we loop over all the facets we found and locate it’s local index in the cell it is associated to.
cell_facet_pairs = np.empty((len(set_of_facets), 2), dtype=np.int32)
for i, facet in enumerate(set_of_facets):
cells = f_to_c.links(facet)
assert len(cells) == 1, "Cell is connected to more than one facet"
facets = c_to_f.links(cells[0])
facet_index = np.flatnonzero(facets == facet)
assert len(facet_index) == 1, "Facet is not connected to cell"
cell_facet_pairs[i] = (cells[0], facet_index[0])
cell_facet_pairs=array([[ 30, 4],
[ 44, 4],
[ 49, 4],
[ 65, 4],
[ 60, 4],
[ 81, 4],
[ 69, 4],
[ 85, 4],
[ 76, 4],
[ 95, 4],
[ 99, 4],
[ 88, 4],
[102, 4],
[ 90, 4],
[105, 4],
[109, 4],
[112, 4],
[104, 4],
[114, 4],
[115, 4],
[118, 4],
[120, 4],
[121, 4],
[123, 4],
[124, 4]], dtype=int32)
We can pass these pairs to the dolfinx.fem.Expression to evaluate
the heat flux on the boundary of the domain.
As before, we can get the coordinates of the points in the physical cell
with ufl.SpatialCoordinate
facet_midpoint = np.array([[0.5, 0.5]], dtype=np.float64)
facet_x = dolfinx.fem.Expression(ufl.SpatialCoordinate(mesh), facet_midpoint)
heat_flux_expr = dolfinx.fem.Expression(heat_flux, facet_midpoint)
coordinates = facet_x.eval(mesh, cell_facet_pairs)
flux_values = heat_flux_expr.eval(mesh, cell_facet_pairs)
coordinate=array([[0.1, 1. , 0.1]]), flux=array([4.1]) exact_flux=[4.1]
coordinate=array([[0.3, 1. , 0.1]]), flux=array([4.1]) exact_flux=[4.1]
coordinate=array([[0.1, 1. , 0.3]]), flux=array([4.3]) exact_flux=[4.3]
coordinate=array([[0.3, 1. , 0.3]]), flux=array([4.3]) exact_flux=[4.3]
coordinate=array([[0.5, 1. , 0.1]]), flux=array([4.1]) exact_flux=[4.1]
coordinate=array([[0.5, 1. , 0.3]]), flux=array([4.3]) exact_flux=[4.3]
coordinate=array([[0.1, 1. , 0.5]]), flux=array([4.5]) exact_flux=[4.5]
coordinate=array([[0.3, 1. , 0.5]]), flux=array([4.5]) exact_flux=[4.5]
coordinate=array([[0.7, 1. , 0.1]]), flux=array([4.1]) exact_flux=[4.1]
coordinate=array([[0.7, 1. , 0.3]]), flux=array([4.3]) exact_flux=[4.3]
coordinate=array([[0.5, 1. , 0.5]]), flux=array([4.5]) exact_flux=[4.5]
coordinate=array([[0.1, 1. , 0.7]]), flux=array([4.7]) exact_flux=[4.7]
coordinate=array([[0.3, 1. , 0.7]]), flux=array([4.7]) exact_flux=[4.7]
coordinate=array([[0.9, 1. , 0.1]]), flux=array([4.1]) exact_flux=[4.1]
coordinate=array([[0.9, 1. , 0.3]]), flux=array([4.3]) exact_flux=[4.3]
coordinate=array([[0.7, 1. , 0.5]]), flux=array([4.5]) exact_flux=[4.5]
coordinate=array([[0.5, 1. , 0.7]]), flux=array([4.7]) exact_flux=[4.7]
coordinate=array([[0.1, 1. , 0.9]]), flux=array([4.9]) exact_flux=[4.9]
coordinate=array([[0.3, 1. , 0.9]]), flux=array([4.9]) exact_flux=[4.9]
coordinate=array([[0.9, 1. , 0.5]]), flux=array([4.5]) exact_flux=[4.5]
coordinate=array([[0.7, 1. , 0.7]]), flux=array([4.7]) exact_flux=[4.7]
coordinate=array([[0.5, 1. , 0.9]]), flux=array([4.9]) exact_flux=[4.9]
coordinate=array([[0.9, 1. , 0.7]]), flux=array([4.7]) exact_flux=[4.7]
coordinate=array([[0.7, 1. , 0.9]]), flux=array([4.9]) exact_flux=[4.9]
coordinate=array([[0.9, 1. , 0.9]]), flux=array([4.9]) exact_flux=[4.9]